 When the ‘one per cent’ truly matters.
When the ‘one per cent’ truly matters.
We’ve all heard that human and chimpanzee genomes are about ninety-nine per cent similar, and many of us have questioned: how could this tiny difference contribute to such drastic differences between the two species? Furthermore, what parts of the human genome are responsible for such differences?
In 2003, after the Human Genome Project was completed, it was found that functional genes (ones that encode protein sequences) constituted only about two per cent of our genome, with the other ninety-eight per cent having no apparent function and thus labeled ‘junk DNA’. However, with time, new evidence pointed out that many non-coding parts of our genome perform regulatory functions. In other words, many non-coding parts of our DNA regulate the expression of other genes through various ways like reducing, enhancing, and silencing it. Furthermore, many of the non-coding sequences turned out to be crucial developmental regulators, thus, being vitally important.
Research estimates that about five to ten per cent of human genome is conserved across mammals, with the majority of it not coding for any proteins. This data argues that the non-coding portion of the human genome therefore contains more functionally constrained DNA than the coding portion. Thus the non-coding portion of the human genome is a larger potential target for evolutionary change. Furthermore, the sequence of protein changes that occurred during human evolution is too small to account for all of the human-specific traits. Evidently, human-specific phenotypes (the composite of an organism’s observable characteristics or traits) must have resulted due to changes in such non-coding regulatory sequences.
How does one determine what exact human-specific changes in the non-coding part of the genome contribute to our unique traits? Thankfully, rapid progress in genome sequencing techniques have allowed multiple vertebrate genomes such as mouse, rat, chimpanzee, zebrafish, amongst others, to be sequenced and mapped, allowing a detailed comparison of those with the human genome. We can identify the regions in human non-coding DNA that are evolving faster than predicted. In other words, the parts of the human non-coding genome that are experiencing faster mutation rates than the analogous parts in other related animals.
Next, the researchers used an experimental design to see whether the human variant of a non-coding sequence of interest has any regulatory role in development. This is done by creating transgenic mice containing human-specific regions of non-coding DNA of interest instead of their native ones. The procedure is repeated to create a transgenic mouse with our ‘ancestral’ form of the non-coding sequence from the chimpanzee. The expression of specific genes in mice embryos containing human and chimpanzee genome sequences can then be compared by using specially constructed ‘reporter genes’ that visualize the expression of the specified gene. If a human variant of such non-coding DNA sequences showed reproducible gene expression different from the related chimpanzee sequence, it can be an indicator of regulatory functions.
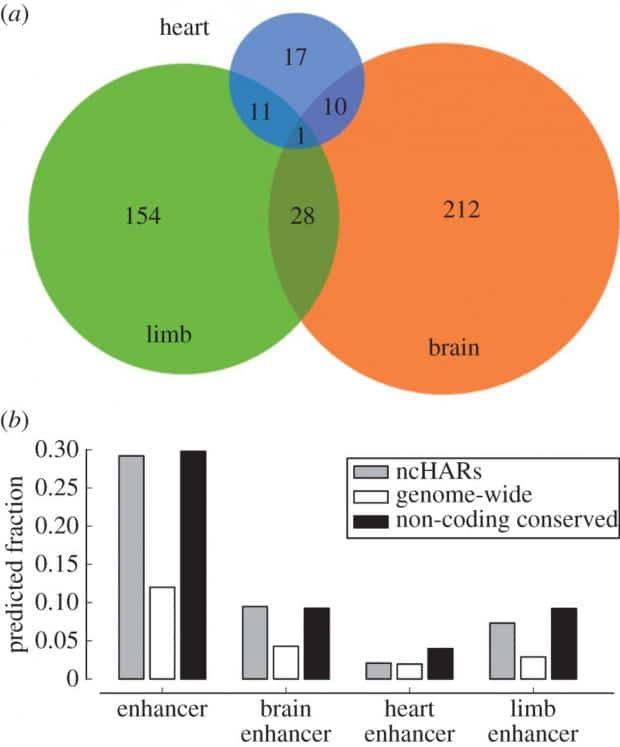
Another approach is to use statistical analysis to predict what parts of the human genome might have such regulatory functions (i.e. enhancer activity) by analyzing their proximity to genes and many other factors involved. A recent study by John A. Capra had predicted that about 773 non-coding human-specific regions of genome with accelerated mutation rates are to be developmental enhancers. Interestingly, the majority of them have been predicted to enhance and regulate brain and limb development—perhaps the most important human traits.





